Cre-loxp system and Viral vector (AAV and adenovirus)-based Cre tools (AAV-Cre and Ad-Cre)
Content index
Product list: Cre/loxP tools in AAV vector(AAV-Cre), adenoviral vector(Ad-Cre) and lentiviral vector(Lv-Cre)
| Cat No. | Pre-made Cre | Viral vector | Protomer | Promoter Tissue Specificity |
Promoter Tissue Specificity characteristics |
| GMV-AdCre-01 | Ad-CMV-Cre | Adenovirus | CMV | Total | Total |
| GMV-AdCre-02 | Ad-CMV-Cre-GFP | CMV | Total | Total | |
| GMV-AAV9Cre-01 | AAV9-CMV-Cre | AAV GeneMedi offer multiple serotypes of AAV-Cre loxP system for your option:
· AAV1-Cre · AAV2-Cre · AAV2 variant (Y444F)-Cre · AAV2 variant (Y272F, Y444F, Y500F, Y730F)-Cre · AAV2 variant (Y444F, Y730F, Y500F, Y272F, Y704F, Y252F)-Cre · AAV2 variant(AAV2.7m8)-Cre · AAV5-Cre · AAV6-Cre · AAV8-Cre · AAV8-1m-Cre · AAV8-2m-Cre · AAV8 variant (Y733F, Y447F, Y275)-Cre · AAV9-Cre · AAV-Rh.10-Cre · AAV-DJ-Cre · AAV-DJ/8-Cre · AAV-Retro (Retrograde)-Cre · AAV-PHP.B-Cre · AAV9-PHP.A-Cre · AAV-PHP.eB-Cre · AAV-PHP.S-Cre | CMV | Total | Total |
| GMV-AAV9Cre-02 | AAV9-CMV-Cre-ZsGreen | CMV | Total | Total | |
| GMV-AAV9Cre-03 | AAV9-CAG-Cre | CAG | Total | Total | |
| GMV-AAV9Cre-04 | AAV9-CAG-Cre-ZsGreen | CAG | Total | Total | |
| GMV-AAV9Cre-05 | AAV9-Syn-Cre-EGFP | synapsin (SYN) | Neuro | Neuron-specific promoter | |
| GMV-AAV9Cre-06 | AAV9-Syn-Cre | synapsin (SYN) | Neuro | Neuron-specific promoter | |
| GMV-AAV9Cre-07 | AAV9-CaMKII-Cre-EGFP | CaMKII | Neuro | Forebrain glutamate neuron-specific promoter | |
| GMV-AAV9Cre-08 | AAV9-CaMKII-Cre | CaMKII | Neuro | Forebrain glutamate neuron-specific promoter | |
| GMV-AAV9Cre-09 | AAV9-GFAP-Cre-EGFP | GFAP | Neuro | Astrocyte specific promoter | |
| GMV-AAV9Cre-10 | AAV9-GFAP-Cre | GFAP | Neuro | Astrocyte specific promoter | |
| GMV-AAV9Cre-11 | AAV9-cTNT-Cre | cTnT | heart | Cardiomyocyte specific promoter | |
| GMV-AAV9Cre-12 | AAV9-cTNT-Cre-EGFP | cTnT | heart | Cardiomyocyte specific promoter | |
| GMV-AAV9Cre-13 | AAV9-TBG-Cre | TBG | liver | Hepatic specific promoter | |
| GMV-AAV9Cre-14 | AAV9-TBG-Cre-ZsGreen | TBG | liver | Hepatic specific promoter | |
| GMV-AAV9Cre-15 | AAV9-TIE-Cre | TIE | endothelial | Endothelial-specific promoter | |
| GMV-AAV9Cre-16 | AAV9-cd68-Cre | cd68 | Neuro | Microglia-specific promoter | |
| GMV-AAV9Cre-17 | AAV9-c-fos-Cre | c-fos | Neuro | Excitatory neuron promoter | |
| GMV-AAV9Cre-18 | AAV9-mecp2-Cre | mecp2 | Neuro | Shorter neuron-specific promoter | |
| GMV-AAV9Cre-19 | AAV9-MHCK7-Cre | MHCK7 | muscle | Muscle specific promoter | |
| GMV-AAV9Cre-20 | AAV9-Rpe65-Cre | Rpe65 | retina | Retinal specific promoter | |
| GMV-AAV9Cre-21 | AAV9-sm22a-Cre | sm22a | muscle | Smooth muscle specific promoter | |
| GMV-AAV9Cre-22 | AAV9-Spc5-12-Cre | Spc5-12 | muscle | Myocyte specific promoter | |
| GMV-AAV9Cre-23 | AAV9-Ta1-Cre | Tal | Neuro | Early neuron-specific promoter | |
| GMV-LvCre-01 | Lv-CMV-Cre | Lentivirus | CMV | Total | Total |
| GMV-LvCre-02 | Lv-CMV-Cre-zsgreen | CMV | Total | Total |
Abstract
Cre-loxP system is widely used in the field of biosciences, especially in the generation of genetically engineered mice (knockout or overexpression), enabling researchers to study the function of gene of interest (GOI). To date, numerous systems, derived from the basic Cre-loxP system, have been developed to precisely control gene expression at desired time or cells. Here, we briefly introduce Cre-loxP system, and give a summary about its applications, especially viral vectors-mediated Cre expression (such as AAV-Cre or Ad-Cre) for in vitro or in vivo studies.
1. Introduction of Cre-loxP system
As a powerful gene editing technology, Cre (cyclization recombinase) was first discovered in bacteriophage P1 to recognize and bind to the specific DNA sequences, named as loxP (locus of crossing over, x, P1) site and thus mediate site-specific rearrangement of target DNA sequences between two loxP sites (Fig. 1A), which can be located in the same or separate DNA fragments [1]. In this recombination process via Cre-loxP system, there were three rearrangement outcomes based on the orientation and location of the two loxP sites, which were shown in Fig. 1: ① gene deletion would happen, if the two loxP sites locate in the same DNA strand as well as the same orientation (Fig. 1B); ② gene inversion would happen, if the two loxP sites locate in the same DNA strand but in opposite orientations (Fig. 1C); ③ sequence translocation would happen, if the sites are in separate DNA molecules (Fig. 1D) [2, 3].

Figure 1. Site-specific recombination via Cre-loxP system. (A) Close-up of Cre recombinase-mediated recombination between two 34 bp loxP sites, which consists of two 13 bp inverted and palindromic repeats (sequences in red or purple) and 8 bp core sequences (sequences in blue, “N” indicates the bases may vary from the canonical sequence of GCATACAT), which determines the orientation of the loxP site [4]. (B-D) Schematic of Cre recombinase mediated target gene deletion (B), inversion (C), or translocation (D).
To date, Cre-loxP system has been widely applied in the generation of gene knockout (KO) mice [5] (Fig. 2), especially the inducible tissue-specific gene KO mice [6]. First, loxP sites should be introduced into the introns to flank an essential exon of targeted DNA (floxed DNA) in murine embryonic stem cells (mESCs) through homologous recombination, which will not affect the expression level of target gene. Then, mESCs need to be injected into a pseudopregnant female mouse blastocyst to develop into a whole mouse carrying the desired targeted mutation. After breeding with the mouse containing Cre gene, the heterozygous mice with target gene excision will be obtained (Fig. 2).

Figure 2. Generation schematics of progeny mice with gene of interest (GOI) mutation via Cre-loxP technology. In principle, once one mouse carrying floxed DNA breeds with another mouse carrying Cre gene, their offspring will have both flanked DNA and Cre. Thus, Cre recombinase will excises floxed loci and inactivates GOI [7]. GOI, gene of interest.
2. Application of inducible tissue-specific KO via Cre-loxP system
To figure out the gene regulation and function more precisely, several sophisticated inducible systems were introduced to control the expression of Cre at specific time (temporal) and tissue (spatial) in vivo or in vitro [7]. There are several common inducible systems via cell-specific regulatory elements (such as promoters or enhancers), exogenous inducers (such as tamoxifen (tam) or tetracycline (tet)) [4, 7-10], and viral vectors (such as adeno-associated viral (AAV) vectors) [11].
1) Inducible-tissue specific Cre-loxP system for in vivo and in vitro study
a) Tissue-specific promoter driven Cre-loxP system for conditional knock-out (KO)
To achieve the conditional knock-out (KO) of gene of interest (GOI) in specific tissue or organ, cell-specific promoter/enhancer driven Cre lines are used to breed with GOI-loxP lines. Their offspring will get both cell-specific driven Cre and floxed DNA (GOI), resulting in tissue-specific GOI excision (Fig. 3). A great number of cell-specific promoters or enhancers have been developed, such as Lyz2 driven Cre specific in macrophage, and CD45 driven Cre specific in hematopoietic tissues. Some examples are listed in the following Table 1.
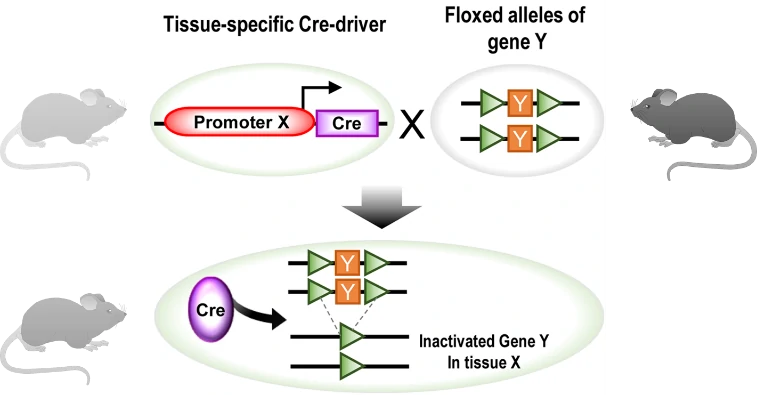
Figure 3. Generation schematics for conditional knock-out (KO) using tissue-specific Cre/loxP tool. In principle, one mouse with tissue (X)-specific driven Cre gene breeds with another mouse with floxed GOI, producing offspring carrying floxed GOI Y and Cre recombinase in tissue X. Thus, GOI Y would be excised by Cre in tissue X [7]. GOI, gene of interest.
| Tissue/organ | cell | Promoter/enhancer | Reference |
| Cerebrum | Neuronal and glia cell precursors | Aldh1l1 | [12] |
| Cerebrum | CaMIIα | [13] | |
| GABAergic interneurons | Dlx1 | [14] | |
| GABAergic neurons | Dlx5/6 | [15-17] | |
| DRG | Gad2 | [18] | |
| Astrocyte | GFAP | [19, 20] | |
| CA3 pyramidal cells | Grik4 | [21] | |
| Arcuate nucleus | Lepr | [22] | |
| Neuronal and glia cell precursors | Nes | [23] | |
| DRG neurons | nNOS | [18] | |
| Retinal Müller glial cells | Pdgfrα | [24] | |
| Myelinating cells | PLP1 | [25] | |
| DRG neurons | Pv (Pvalb) | [18] | |
| GABAergic neurons | Slc17a6 | [26] | |
| GABAergic neurons | Sst, Vip | [17] | |
| Cerebellum | Cerebellar Purkinje cells | Pcp2 | [27] |
| Brain stem | Dopamine and serotonin neurons | Slc6a3 (DAT) | [28] |
| Serotonin neurons | ePet (Fev) | [29] | |
| Vagal sensory neurons | Npy2r | [30] | |
| Spinal cord | Mechanosensory dorsal horn | Cdh3, Htr6 | [31] |
| Skin | Basal layer of the epidermis | Krt5 | [32] |
| Epidermis keratinocytes | Krt10 | [33] | |
| Epidermal progenitors | Krt18 | [34] | |
| Immune system | Macrophage | Lyz2 | [35] |
| Dendritic cells | CD11c (Itgax) | [36] | |
| Mast cells | CMA1 | [37] | |
| T cells | CD2 | [38] | |
| CD4 | [39] | ||
| CD8a | [40] | ||
| Foxp3 | [41] | ||
| Lck | [42] | ||
| OX40 | [43] | ||
| B cells | CD19 | [44, 45] | |
| Hematopoietic cells | CD15 | [46] | |
| Musculoskeletal | Osteochondro progenitors | Twist2 (Dermol) | [47, 48] |
| Prrx1 | [49] | ||
| Osteoblasts | BGLOP | [50] | |
| Osteocytes | Dmp1 | [51] | |
| Osteoclasts | Ctsk | [52] | |
| Cardiomyocytes | NKX2.5 | [53] | |
| Muscle | ACTA1 (HSA) | [54] | |
| Digestive system | Stomach | ATP4b | [55] |
| Intestine | Car1 | [56] | |
| Intestine | Vil1 | [57] | |
| Liver | Alb | [58, 59] | |
| PanCreas | Ghrl | [60] | |
| Ins1 (MIP) | [61] | ||
| Ins2 (RIP) | [62] | ||
| Ptf1a | [63] | ||
| Urogenital system | Bladder | Upk2 | [64] |
| Kidney | Aqp2 | [65] | |
| Gdnf | [66] | ||
| Ggt1 | [67] | ||
| Ovary | Gdf9 | [68] | |
| Zp3 | [69] | ||
| Testis | Amh | [70] | |
| Hspa2 | [71] |
One inducible system is tamoxifen-inducible Cre-loxP system (Fig. 4A). When ubiquitous or tissue-specific promoter driven Cre is fused with the estrogen receptor containing a mutated ligand binding domain (ER-LBD), which is normally bound by heat shock protein 90 (HSP90), Cre-ER-HSP90 complex would be prevented from entering the nucleus. Once bound by the hormone (such as estrogen) or synthetic analogs (such as tamoxifen, T, or 4-hydoxytamoxifen, 4-OHT), ER will be released from HSP90. This enables the nuclear translocation of Cre-ERT complex and the following interaction between Cre and loxP sites [8, 32].
Another inducible system is tetracycline inducible Cre-loxP system, also known as doxycycline (dox; a tetracycline derivative)-inducible Cre system. This system has two mode: tet-on (Fig. 4B) and tet-off (Fig. 4C), responsible for the activation and inactivation of Cre gene, respectively [72, 73]. There are three elements orchestrating to control Cre expression in this system: reverse tetracycline-controlled transactivator (rtTA), tetracycline-controlled transactivator (tTA) and tetracycline responsive element (TRE), also named as a tetracycline operon (TetO). In tet-on system, ubiquitous or tissue-specific promoter driven rtTA binds to dox to turn on the transcription of Cre gene (Fig. 4B). On the contrary, in tet-off system, tTA binds to dox to turn off the transcription of Cre gene (Fig. 4C).

Figure 4. Operational principles of inducible tissue-specific Cre-loxP system [7]. (A) Tamoxifen-inducible Cre-loxP system with estrogen receptor (ER) fused to Cre (CreER). When tamoxifen is supplemented to bind to ER, Cre will translocate into nucleus to mediate the excision target gene Y. (B) Tetracycline (Tet)-on Cre-loxP system. Without dox, inactivated rtTA cannot activate the tetO7 (7 repeats of a 19 nuclotide tetO minimal promoter) for Cre expression. Upon the supplementation of dox, activated rtTA binds to tetO7 to initiate the transcription of Cre gene. (C) Tet-off Cre-loxP system. In the absence of dox, tTA is able to bind to tetO7 to activate Cre expression. Following the administration of dox, tTA will be silenced as well as lose the ability to bind to tetO7 and activate Cre transcription.
So far, a huge number of Cre transgenic lines have been established to combine with GOI loxP lines to generate conditional knockout mice or cell lines at the desired period and tissues in vitro or in vivo. Given that it is time and labor consuming to use Cre transgenic mice lines, viral vectors are convenient and versatile tools for the expression of Cre in certain cells or tissues, such as adeno-associated viral (AAV) vectors, as well as cell lines, such as adenovirus (Ad).
2) Tissue-specific AAV-Cre for in vivo study
As the most excellent gene therapy vector, recombinant AAV can mediate long-term stable expression of target genes in vitro/vivo with broad range of host and low immunity. Over the past decades, numerous AAV serotypes have been discovered or developed, and each serotype has its special tropism. For instance, AAV2 can moderately transduce several tissue types, including central nervous system, liver, muscle, and lung. Similarly within the CNS, AAV1 and AAV5 show higher transduction frequencies than AAV2 in all injected regions [74, 75], while AAV4 just appears to transduce specific cell types, such as the astrocytes and ependyma in the subventricular zone [76]. In skeletal muscle cells, AAV1, AAV6 and AAV7 is reported to present very high transduction rate [77-80], while AAV8 exhibits transduction ability with super efficiency not only in hepatocytes but also in other organs [78, 81, 82]. In addition to skeletal muscle cells, AAV6 has been also reported to mediate more efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 [81], which may supply AAV6 with more significant advantages over AAV2 for gene therapy of lung diseases like cystic fibrosis considering the lower immunogenicity of AAV6 than of AAV2 [78, 82].
GeneMedi holds the expertise at AAV production, you can find more information and protocols about AAV on this website: https://www.genemedi.net/i/aav-packaging.
Combined with Cre-loxP system, AAV containing Cre (AAV-Cre) can help mediate GOI conditional KO in specific tissues or organs based on its special tissue trophism (Fig. 5). This technology has been widely applied into several research fields, especially for the gene function studies in neurosciences. Some of examples are listed in Table 2.

Figure 5. Generation schematics for AAV-mediated tissue-specific GOI deletion. About two weeks post-infection of AAV-Cre viruses, the transgenic mice with floxed DNA will express Cre in specific tissue based on the AAV tropism. Then, in this specific tissue, Cre recombinase will bind and excise the loxP sites, thus inactivate the GOI.
| AAV serotype | Tissue/organ | Delivery route | Gene | Reference |
| AAV9 | Hippocampus or central amygdala | Stereotaxic microinjection | IRs and IGF1Rs | [11] |
| AAV9 | Astrocyte | In vitro transduction | IGFR | [83] |
| AAV2/5 | Large DRG neurons | Intrathecal injection | Na(V)1.6 | [84] |
| AAV9 | Spinal dorsal horn neurons | Intrathecal injection | SIRT1 | [85] |
| AAV2 | Cortical neurons | Unilateral intracortical injections | PTEN | [86] |
| AAV5 | Forebrain excitatory neurons | Stereotaxic microinjection | SIRT1 | [87] |
| AAV2/1 | Muscle | Intramuscular injection | Mtm1 | [88] |
| AAV2 | Ventral tegmental area, VTA | Intra-VTA infusion | Gabrd | [89] |
| AAV8 | Sensorimotor cortex, hippocampus, and spinal cord | Intra-sensorimotor cortex injection | PTEN | [90] |
| AAV2/5 | mesolimbic Ventral tegmental area (VTA), NAc core and central amygdala (CeA) | Intracerebroventricular (ICV) injection | Lepr | [91] |
| AAV9 | Dentate gyrus (DG) | Intra-DG injection | Shh | [92] |
| AAV8 | Liver | Lateral tail vein injection | HKDC1 | [93] |
| AAV1 | Hypothalamus | Stereotaxic microinjection | AC3 | [94] |
Additionally, tissue-specific promoter is also used to drive Cre expression to strengthen the tissue specificity of GOI knockout. Several tissue-specific promoters, within the loading capacity of AAV vectors, are listed in the following table 3.
| Tissue/organ | Promoter | Specificity | Promoter size | Species |
| Neurocyte | hSyn | Neuron | 0.5kb | human |
| mecp2 | Short neuron | 0.2kb | mouse | |
| TUBA1A | Developing neuron | 1.2kb | human | |
| c-fos | Excitatory neuron | 1.7kb | mouse | |
| CaMKIIa | Glutamate neurons in the forebrain | 1.1kb | mouse | |
| hVGAT | GABAergic interneurons | 1.8kb | human | |
| Slc6a3 | Dopamine and serotonin neurons | 1.2kb | mouse | |
| gfaABC1D | Astrocyte | 0.9kb | human | |
| Iba1 | Keratinocyte | 1.5kb | mouse | |
| CNP | Schwann cell | 1.5kb | mouse | |
| Muscle | cTNT | Cardiomyocyte | 0.7kb | chicken |
| mNKX2.5 | Cardiomyocyte | 1.2kb | mouse | |
| SM22a | Smooth muscle | 0.5kb | mouse | |
| MCK | Muscle | 1.3kb | mouse | |
| MYOG | Myoblast | 1.4kb | mouse | |
| ACTA1 | Myocyte | 1.2kb | mouse | |
| Skeleton | COL2A1 | Chondrocyte | 1.2kb | mouse |
| mRUNX2 | Osteoblast | 1kb | mouse | |
| Lung | SP-C | Lung epithelial cells | 0.3kb | mouse |
| Liver | TBG | Liver cells | 0.4kb | human |
| PanCreas | PDX1 | PanCreas cells | 1.2kb | mouse |
| Fat | FABP4 | Fat cells | 1.6kb | mouse |
| Endothelial | TIE | Endothelial cells | 1.2kb | mouse |
| Corneum | K14 | Keratinocyte | 1.5kb | mouse |
| Retina | rpe65 | Retinal cells | 0.7kb | mouse |
GeneMedi offers kinds of pre-made AAV-Cre particles in different serotypes driven by optional promoters. Click here for more information about GeneMedi’s ready-to-use AAV-Cre:https://www.genemedi.net/i/Cre-products
3) Tissue/cell-specific Ad-Cre for in vivo or in vitro study
Based on human adenovirus type 5 (Ad5), recombinant adenovirus (Ad) a replication-defective adenoviral vector system, is widely used for gene delivery in most cell types [95], and Genemedi got a rich experience in adenovirus packaging, you could find more information on https://www.genemedi.net/i/adenovirus-packaging.
Combined with Cre-loxP system and cell-specific promoter driven Cre, Adenoviral vector can also help mediate GOI conditional KO in vivo. Besides, Adenoviral vector containing Cre (Ad-Cre) also exhibits great advantages and high efficiency for the in vitro studies, especially for the transduction of primary cells [96], which are commonly difficult to be transduced (Fig. 5). As a promising and efficient tool, Ad-Cre system has been extensively used into the research field of cell lineage tracing and fate mapping. Some of examples are listed in Table 4.
| Promoter | Tissue/organ/cell | Delivery route | Gene | Reference |
| Keratin 8/ Keratin 5 | Mammary epithelial cells | Intraductal injection | YFP | [97] |
| Keratin 8/ Keratin 14 | Mammary epithelial cells | Intraductal injection | YFP | [98] |
| —— | Leydig cells | Intratesticular injection | Gata4 and Gata6 | [99] |
| CMV | Subfornical organ-hypothalamic-hypophysial and brain stem-parabrachial axes | Stereotaxic microinjections | β-galactosidase (β-gal) | [96] |
| CMV | Vasopressin-synthesizing magnocellular neurons | Retrograde transport | β-galactosidase (β-gal) | [96] |
| CMV | Primary neuronal cells | In vitro inoculation | β-galactosidase (β-gal) | [96] |
| —— | PASMCs | In vitro inoculation | ALK2 | [100] |
| —— | Muscle | Retropopliteal injection | ALK2 | [100] |
| CAG | HepG2 cell line | In vitro inoculation | SV40Tag | [101] |
| —— | Primary osteoblasts | In vitro inoculation | IGF-IR | [102] |
| —— | Mesangial cells | In vitro inoculation | EP4 | [103] |
| —— | Primary calvarial cells | In vitro inoculation | IGF-IR | [104] |
GeneMedi offers kinds of pre-made Adenovirus-Cre (Ad5-Cre, Ad-Cre) particle driven by optional promoters. Click here for more information about GeneMedi’s ready-to-use Adenovirus-Cre (Ad5-Cre, Ad-Cre):https://www.genemedi.net/i/Cre-products
3. Cre-loxP induced overexpression: Cre-DIO system
Besides GOI deletion, Cre-loxP system also shows great advantages in GOI overexpression, such as Cre-dependent GOI expression, and Cre-DIO (double-floxed inverse open reading frame) system. 1) Cre-dependent GOI expression: a stop codon between two loxP sites (loxP-stop-loxP, LSL, floxed stop) was placed at the upstream of GOI, thus preventing the expression GOI. Once Cre is supplemented, the stop codon and one lox site would be excised, and the GOI expression would be activated [105]. 2) Considering that the three modes of Cre-loxP technology are all reversible, Cre-DIO system were created to induce inverse gene expression utilizing two pairs of lox sites [106, 107].
Principle of Cre-DIO
Although Cre-loxP tool is widespread in biological sciences, it is really restrictive to achieve more than one recombination event in a cell. Therefore, numerous loxP mutant versions have been created, with the variation from the unique 8 bp core asymmetric spacer "NNNTANNN", such as loxN (GtATACcT), lox2272 (GgATACtT), lox5171 (ATGTGTaC), loxM2 (AgaaAcca), loxM3 (taaTACCA), loxM11 (AGATAGAA), and lox511 (GtATACAT). These lox variants only proceed recombination with the same type of lox sites, with no ability to interact with the other types. In Cre-DIO system, there are two pairs of lox sites (lox site 1 and lox site 2) flanking inverted GOI and reporter gene. In the absence of Cre, the inverted GOI and reporter are inactivated. Upon administration of Cre, the two pairs of lox sites will undergo recombination, resulting in the expression of GOI and reporter gene. Simultaneously, the cells with recombination interaction are labeled by the reporter gene (Fig. 6). The Cre-DIO tool utilizes initially inverted GOI to truly inactivate GOI, and activates GOI after a series of recombination interactions. Compared to the traditional overexpression strategy via floxed stop, this technology is much more effective to avoid the transcriptional leakage when the stop codon fails to totally suppress the transcription of GOI.

Figure 6. Principle of Cre-DIO (double-floxed inverse open reading frame) system. Without Cre, the GOI and reporter gene are inverted and silenced. In the presence of Cre, first, the transgene will be reoriented, then one of the two lox sites in each pair will be excised. Finally, the transgene contains only an incompatible pair of lox sites (lox 2-GOI-Reporter-lox 1) that are no longer responsive to Cre. Thus, the GOI and reporter gene will be activated. Lox site 1 and lox site 2 are variants of lox sites.
Tissue-specific Cre mouse + AAV-DIO-GOI for tissue-specific gene overexpression
To facilitate the use of Cre-DIO system, AAV vectors have been introduced to tissue-specific Cre mouse to mediate tissue-specific gene overexpression. Without injection of AAV vector carrying DOI-GOI system (AAV-DIO-GOI), tissue X-specific Cre mouse cannot express GOI and reporter gene in tissue X. After injection of AAV-DIO-GOI into tissue X, Cre recombinase will interact with lox sites, thus activating the expression of GOI and reporter. Synchronously, the tissue X will be labeled by the reporter gene (Fig. 7). This system utilizes high cell-specific promoter to drive Cre recombinase expression and a strong promoter to drive GOI expression, which ideally occur just at the appropriate time point and specific cells. As a powerful system with high spatiotemporal fidelity, AAV-DIO exhibits great advantages and potential in the study of neural circuits and networks, behavior, specific cell populations (lineage tracing), animal models of disease, as well as in high-throughput ex vivo researches.

Figure 7. Principle of tissue-specific gene overexpression via AAV-DIO-GOI with tissue-specific Cre mouse. Without AAV-DOI-GOI, the GOI and reporter gene do not express in tissue X. Upon AAV-DOI-GOI administration into the mouse with tissue X-specific Cre, the GOI and reporter gene will be activated in tissue X and the tissue X will be labeled by reporter gene (green). Lox site 1 and lox site 2 are variants of lox sites.
AAV-DIO in neuroscience
In the research field of neuroscience, AAV-DIO shows outstanding spatiotemporal fidelity and is perfectly suitable for examining and manipulating neuronal function in vivo, such as studies in neural circuits and networks, behavior and so on. Optogenetics integrates optics with genetic engineering to measure or manipulate the biomolecular processes, especially in neuroscience [108]. By delivering optical signals with specific wavelength to precisely detect, measure, or control biological processes in living tissues, optogenetics opens up new landscapes for the study of biology, both in health and disease [109]. With the combination of AAV-DIO and optogenetics, researchers control and monitor the activities of individual neurons, and precisely measure the manipulation effects in real-time. For example, in Fig. 8, with AAV-DIO system, the neuron X specifically express opsin (such ChR2) and is labeled by the reporter gene (such as mCherry), but remains silenced. When exposed to light in correct wavelength, neuron X will be activated (Fig. 8) [110].

Figure 8. Principle of optogenetics tool in combination with AAV-DIO. With no light in specific wavelength matched to opsin, such as ChR2, ChR2 gene and the reporter gene is specifically expressed in neurons (location in red). Once exposed to light with the matched wavelength, ChR2 cation channels will be open and Na+ flow into neural cells. Thus, the neuron will be activated.
Genemedi possesses a complete library of optogenetics components into serotypes of AAV vector, which is listed in table 5. You can find more information on this website: https://www.genemedi.net/i/aav-optogenetics.
| AAV Plasmids | Promoter and Expression Characteristics | Fluorescent Protein Label | Cre dependent | Optogenetics effect |
| pAAV-CAG-DIO-eNpHR3.0-EYFP | CAG general and strong expression | EYFP | yes | Inhibition |
| pAAV-CAG -DIO-hChR2(H134R)-mCherry | mCherry | yes | activation | |
| pAAV-CAG-DIO-ArchT-EYFP | EYFP | yes | inhibition | |
| pAAV-CAG -DIO-C1V1 (t/t)-TS-mCherry | mCherry | yes | activation | |
| pAAV-CAG-DIO-Arch3.0-EYFP | EYFP | yes | inhibition | |
| pAAV- CAG -DIO-hCHETA-EYFP | EYFP | yes | activation | |
| pAAV-CMV-DIO-eNpHR3.0-EYFP | CMV(general and strong expression) | EYFP | yes | inhibition |
| pAAV-CMV-DIO-hChR2(H134R)-mCherry | mCherry | yes | activation | |
| pAAV-CMV-DIO-ArchT-EYFP | EYFP | yes | inhibition | |
| pAAV-CMV-DIO-C1V1 (t/t)-TS-mCherry | mCherry | yes | activation | |
| pAAV-CMV-DIO-Arch3.0-EYFP | EYFP | yes | inhibition | |
| pAAV-CMV-DIO-hCHETA-EYFP | EYFP | yes | activation | |
| pAAV-hSyn-eNpHR3.0-EYFP | hSyn neuron specific expression | EYFP | No | inhibition |
| pAAV-hSyn-hChR2(H134R)-mCherry | mCherry | No | activation | |
| pAAV-hSyn-ArchT-EYFP | EYFP | No | inhibition | |
| pAAV-hSyn-C1V1-(t/t)-TS-mCherry | mCherry | No | activation | |
| pAAV-hSyn-Arch3.0-EYFP | EYFP | No | inhibition | |
| pAAV-hSyn-DIO-hCHETA-EYFP | EYFP | yes | activation | |
| pAAV-GFAP-eNpHR3.0-EYFP | GFAP astrocyte specific expression | EYFP | No | inhibition |
| pAAV-GFAP-hChR2(H134R)-mCherry | mCherry | No | activation | |
| pAAV-GFAP-ArchT-EYFP | EYFP | No | inhibition | |
| pAAV-GFAP-C1V1 (t/t)-TS-mCherry | mCherry | No | activation | |
| pAAV-GFAP-Arch3.0-EYFP | EYFP | No | inhibition | |
| pAAV-GFAP-hCHETA-EYFP | EYFP | No | activation | |
| pAAV-CaMKII-eNpHR3.0-EYFP | CaMKII neuron specific expression | EYFP | No | inhibition |
| pAAV-CaMKII-hChR2(H134R)-mCherry | mCherry | No | activation | |
| pAAV-CaMKII-ArchT-EYFP | EYFP | No | inhibition | |
| pAAV-CaMKII-C1V1 (t/t)-TS-mCherry | mCherry | No | activation | |
| pAAV-CaMKII-Arch3.0-EYFP | EYFP | No | inhibition | |
| pAAV-CaMKII-hCHETA-EYFP | EYFP | No | activation |
4. Summary
The Cre-loxP system, especially inducible tissue-specific knockout and viral vector-mediated Cre expression, has been highly utilized in genetics and cell biology research. Nevertheless, it seems that Cre-loxP system will continue to be prevalent in current and future research studies. GeneMedi is proficient in viral vector development and offers kinds of viral vector-based Cre tools, we can provide the best services and products if required.
5. References
1.
Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150:467-486.
2.
Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc Natl Acad Sci U S A. 2002;99:4185-4190.
3.
McLellan MA, Rosenthal NA, Pinto AR. Cre-loxP-Mediated Recombination: General Principles and Experimental Considerations. Curr Protoc Mouse Biol. 2017;7:1-12.
4.
Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99-109.
5.
Hall B, Limaye A, Kulkarni AB. Overview: generation of gene knockout mice. Curr Protoc Cell Biol. 2009;Chapter 19:Unit 19 12 19 12 11-17.
6.
Kos CH. Cre/loxP system for generating tissue-specific knockout mouse models. Nutr Rev. 2004;62:243-246.
7.
Kim H, Kim M, Im SK, Fang S. Mouse Cre-LoxP system: general principles to determine tissue-specific roles of target genes. Lab Anim Res. 2018;34:147-159.
8.
Metzger D, Chambon P. Site- and time-specific gene targeting in the mouse. Methods. 2001;24:71-80.
9.
Brocard J, Warot X, Wendling O, Messaddeq N, Vonesch JL, Chambon P, et al. Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc Natl Acad Sci U S A. 1997;94:14559-14563.
10.
Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427-1429.
11.
Soto M, Cai W, Konishi M, Kahn CR. Insulin signaling in the hippocampus and amygdala regulates metabolism and neurobehavior. Proc Natl Acad Sci U S A. 2019;116:6379-6384.
12.
Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817-9823.
13.
Dragatsis I, Zeitlin S. CaMKIIalpha-Cre transgene expression and recombination patterns in the mouse brain. Genesis. 2000;26:133-135.
14.
Potter GB, Petryniak MA, Shevchenko E, McKinsey GL, Ekker M, Rubenstein JL. Generation of Cre-transgenic mice using Dlx1/Dlx2 enhancers and their characterization in GABAergic interneurons. Mol Cell Neurosci. 2009;40:167-186.
15.
Zerucha T, Stuhmer T, Hatch G, Park BK, Long Q, Yu G, et al. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709-7
16.
Monory K, Massa F, Egertova M, Eder M, Blaudzun H, Westenbroek R, et al. The endocannabinoid system controls key epileptogenic circuits in the hippocampus. Neuron. 2006;51:455-466.
17.
Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995-1013.
18.
Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159.
19.
Garcia AD, Doan NB, Imura T, Bush TG, Sofroniew MV. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat Neurosci. 2004;7:1233-1241.
20.
Brenner M, Kisseberth WC, Su Y, Besnard F, Messing A. GFAP promoter directs astrocyte-specific expression in transgenic mice. J Neurosci. 1994;14:1030-1037.
21.
Nakazawa K, Quirk MC, Chitwood RA, Watanabe M, Yeckel MF, Sun LD, et al. Requirement for hippocampal CA3 NMDA receptors in associative memory recall. Science. 2002;297:211-218.
22.
DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, et al. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608-2613.
23.
Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99-103.
24.
Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, et al. The transcriptome of retinal Muller glial cells. J Comp Neurol. 2008;509:225-238.
25.
Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35:63-72.
26.
Vong L, Ye C, Yang Z, Choi B, Chua S, Jr., Lowell BB. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142-154.
27.
Zhang XM, Ng AH, Tanner JA, Wu WT, Copeland NG, Jenkins NA, et al. Highly restricted expression of Cre recombinase in cerebellar Purkinje cells. Genesis. 2004;40:45-51.
28.
Zhuang X, Masson J, Gingrich JA, Rayport S, Hen R. Targeted gene expression in dopamine and serotonin neurons of the mouse brain. J Neurosci Methods. 2005;143:27-32.
29.
Scott MM, Wylie CJ, Lerch JK, Murphy R, Lobur K, Herlitze S, et al. A genetic approach to access serotonin neurons for in vivo and in vitro studies. Proc Natl Acad Sci U S A. 2005;102:16472-16477.
30.
Chang RB, Strochlic DE, Williams EK, Umans BD, Liberles SD. Vagal Sensory Neuron Subtypes that Differentially Control Breathing. Cell. 2015;161:622-633.
31.
Abraira VE, Kuehn ED, Chirila AM, Springel MW, Toliver AA, Zimmerman AL, et al. The Cellular and Synaptic Architecture of the Mechanosensory Dorsal Horn. Cell. 2017;168:295-310 e219.
32.
Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, et al. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER
33.
Calleja C, Messaddeq N, Chapellier B, Yang H, Krezel W, Li M, et al. Genetic and pharmacological evidence that a retinoic acid cannot be the RXR-activating ligand in mouse epidermis keratinocytes. Genes Dev. 2006;20:1525-1538.
34.
Van Keymeulen A, Mascre G, Youseff KK, Harel I, Michaux C, De Geest N, et al. Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J Cell Biol. 2009;187:91-100.
35.
Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265-277.
36.
Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204:1653-1664.
37.
Musch W, Wege AK, Mannel DN, Hehlgans T. Generation and characterization of alpha-chymase-Cre transgenic mice. Genesis. 2008;46:163-166.
38.
Vacchio MS, Wang L, Bouladoux N, Carpenter AC, Xiong Y, Williams LC, et al. A ThPOK-LRF transcriptional node maintains the integrity and effector potential of post-thymic CD4+ T cells. Nat Immunol. 2014;15:947-956.
39.
Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917-929.
40.
Maekawa Y, Minato Y, Ishifune C, Kurihara T, Kitamura A, Kojima H, et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat Immunol. 2008;9:1140-1147.
41.
Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, et al. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667-1671.
42.
Hennet T, Hagen FK, Tabak LA, Marth JD. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc Natl Acad Sci U S A. 1995;92:12070-12074.
43.
Klinger M, Kim JK, Chmura SA, Barczak A, Erle DJ, Killeen N. Thymic OX40 expression discriminates cells undergoing strong responses to selection ligands. J Immunol. 2009;182:4581-4589.
44.
Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317-1318.
45.
Yasuda T, Wirtz T, Zhang B, Wunderlich T, Schmidt-Supprian M, Sommermann T, et al. Studying Epstein-Barr virus pathologies and immune surveillance by reconstructing EBV infection in mice. Cold Spring Harb Symp Quant Biol. 2013;78:259-263.
46.
Yang J, Hills D, Taylor E, Pfeffer K, Ure J, Medvinsky A. Transgenic tools for analysis of the haematopoietic system: knock-in CD45 reporter and deletor mice. J Immunol Methods. 2008;337:81-87.
47.
Sosic D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169-180.
48.
Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063-3074.
49.
Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77-80.
50.
Zhang M, Xuan S, Bouxsein ML, von Stechow D, Akeno N, Faugere MC, et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Che
51.
Lu Y, Xie Y, Zhang S, Dusevich V, Bonewald LF, Feng JQ. DMP1-targeted Cre expression in odontoblasts and osteocytes. J Dent Res. 2007;86:320-325.
52.
Kim JE, Nakashima K, de Crombrugghe B. Transgenic mice expressing a ligand-inducible cre recombinase in osteoblasts and odontoblasts: a new tool to examine physiology and disease of postnatal bone and tooth. Am J Pathol. 2004;165:1875-1882.
53.
Diehl F, Brown MA, van Amerongen MJ, Novoyatleva T, Wietelmann A, Harriss J, et al. Cardiac deletion of Smyd2 is dispensable for mouse heart development. PLoS One. 2010;5:e9748.
54.
Miniou P, Tiziano D, Frugier T, Roblot N, Le Meur M, Melki J. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res. 1999;27:e27.
55.
Syder AJ, Karam SM, Mills JC, Ippolito JE, Ansari HR, Farook V, et al. A transgenic mouse model of metastatic carcinoma involving transdifferentiation of a gastric epithelial lineage progenitor to a neuroendocrine phenotype. Proc Natl Acad
56.
Xue Y, Johnson R, Desmet M, Snyder PW, Fleet JC. Generation of a transgenic mouse for colorectal cancer research with intestinal cre expression limited to the large intestine. Mol Cancer Res. 2010;8:1095-1104.
57.
Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 200
58.
Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, et al. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci U S A. 1999;96:7324-7329.
59.
Stratikopoulos E, Szabolcs M, Dragatsis I, Klinakis A, Efstratiadis A. The hormonal action of IGF1 in postnatal mouse growth. Proc Natl Acad Sci U S A. 2008;105:19378-19383.
60.
Arnes L, Hill JT, Gross S, Magnuson MA, Sussel L. Ghrelin expression in the mouse pancreas defines a unique multipotent progenitor population. PLoS One. 2012;7:e52026.
61.
Thorens B, Tarussio D, Maestro MA, Rovira M, Heikkila E, Ferrer J. Ins1(Cre) knock-in mice for beta cell-specific gene recombination. Diabetologia. 2015;58:558-565.
62.
Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:3
63.
Kopinke D, Brailsford M, Pan FC, Magnuson MA, Wright CV, Murtaugh LC. Ongoing Notch signaling maintains phenotypic fidelity in the adult exocrine pancreas. Dev Biol. 2012;362:57-64.
64.
Ayala de la Pena F, Kanasaki K, Kanasaki M, Tangirala N, Maeda G, Kalluri R. Loss of p53 and acquisition of angiogenic microRNA profile are insufficient to facilitate progression of bladder urothelial carcinoma in situ to invasive carcinoma
65.
Ge Y, Ahn D, Stricklett PK, Hughes AK, Yanagisawa M, Verbalis JG, et al. Collecting duct-specific knockout of endothelin-1 alters vasopressin regulation of urine osmolality. Am J Physiol Renal Physiol. 2005;288:F912-920.
66.
Cebrian C, Asai N, D'Agati V, Costantini F. The number of fetal nephron progenitor cells limits ureteric branching and adult nephron endowment. Cell Rep. 2014;7:127-137.
67.
Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110:341-350.
68.
Lan ZJ, Xu X, Cooney AJ. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol Reprod. 2004;71:1469-1474.
69.
Lewandoski M, Wassarman KM, Martin GR. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol. 1997;7:148-151.
70.
Holdcraft RW, Braun RE. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development. 2004;131:459-467.
71.
Inselman AL, Nakamura N, Brown PR, Willis WD, Goulding EH, Eddy EM. Heat shock protein 2 promoter drives Cre expression in spermatocytes of transgenic mice. Genesis. 2010;48:114-120.
72.
Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, et al. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci U S A. 1996;93:10933-10938.
73.
Pluta K, Luce MJ, Bao L, Agha-Mohammadi S, Reiser J. Tight control of transgene expression by lentivirus vectors containing second-generation tetracycline-responsive promoters. J Gene Med. 2005;7:803-817.
74.
Alisky JM, Hughes SM, Sauter SL, Jolly D, Dubensky TW, Jr., Staber PD, et al. Transduction of murine cerebellar neurons with recombinant FIV and AAV5 vectors. Neuroreport. 2000;11:2669-2673.
75.
Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, et al. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to differ
76.
Davidson BL, Stein CS, Heth JA, Martins I, Kotin RM, Derksen TA, et al. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proceedings of the
77.
Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, et al. Clades of Adeno-associated viruses are widely disseminated in human tissues. Journal of virology. 2004;78:6381-6388.
78.
Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1185
79.
Chao H, Liu Y, Rabinowitz J, Li C, Samulski RJ, Walsh CE. Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Molecular therapy : the journal of the American Society of Gene Therapy. 2
80.
Blankinship MJ, Gregorevic P, Allen JM, Harper SQ, Harper H, Halbert CL, et al. Efficient transduction of skeletal muscle using vectors based on adeno-associated virus serotype 6. Molecular therapy : the journal of the American Society of G
81.
Halbert CL, Allen JM, Miller AD. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. Journal of virology. 2001;75:6615-6624.
82.
Rabinowitz JE, Bowles DE, Faust SM, Ledford JG, Cunningham SE, Samulski RJ. Cross-dressing the virion: the transcapsidation of adeno-associated virus serotypes functionally defines subgroups. Journal of virology. 2004;78:4421-4432.
83.
Logan S, Pharaoh GA, Marlin MC, Masser DR, Matsuzaki S, Wronowski B, et al. Insulin-like growth factor receptor signaling regulates working memory, mitochondrial metabolism, and amyloid-beta uptake in astrocytes. Mol Metab. 2018;9:141-155.
84.
Chen L, Huang J, Zhao P, Persson AK, Dib-Hajj FB, Cheng X, et al. Conditional knockout of NaV1.6 in adult mice ameliorates neuropathic pain. Sci Rep. 2018;8:3845.
85.
Zhang Z, Ding X, Zhou Z, Qiu Z, Shi N, Zhou S, et al. Sirtuin 1 alleviates diabetic neuropathic pain by regulating synaptic plasticity of spinal dorsal horn neurons. Pain. 2019;160:1082-1092.
86.
Gallent EA, Steward O. Neuronal PTEN deletion in adult cortical neurons triggers progressive growth of cell bodies, dendrites, and axons. Exp Neurol. 2018;303:12-28.
87.
Lei Y, Wang J, Wang D, Li C, Liu B, Fang X, et al. SIRT1 in forebrain excitatory neurons produces sexually dimorphic effects on depression-related behaviors and modulates neuronal excitability and synaptic transmission in the medial prefron
88.
Joubert R, Vignaud A, Le M, Moal C, Messaddeq N, Buj-Bello A. Site-specific Mtm1 mutagenesis by an AAV-Cre vector reveals that myotubularin is essential in adult muscle. Hum Mol Genet. 2013;22:1856-1866.
89.
Darnieder LM, Melon LC, Do T, Walton NL, Miczek KA, Maguire JL. Female-specific decreases in alcohol binge-like drinking resulting from GABAA receptor delta-subunit knockdown in the VTA. Sci Rep. 2019;9:8102.
90.
Yang P, Qin Y, Zhang W, Bian Z, Wang R. Sensorimotor Cortex Injection of Adeno-Associated Viral Vector Mediates Knockout of PTEN in Neurons of the Brain and Spinal Cord of Mice. J Mol Neurosci. 2015;57:470-476.
91.
Shen M, Jiang C, Liu P, Wang F, Ma L. Mesolimbic leptin signaling negatively regulates cocaine-conditioned reward. Transl Psychiatry. 2016;6:e972.
92.
Gonzalez-Reyes LE, Chiang CC, Zhang M, Johnson J, Arrillaga-Tamez M, Couturier NH, et al. Sonic Hedgehog is expressed by hilar mossy cells and regulates cellular survival and neurogenesis in the adult hippocampus. Sci Rep. 2019;9:17402.
93.
Khan MW, Priyadarshini M, Cordoba-Chacon J, Becker TC, Layden BT. Hepatic hexokinase domain containing 1 (HKDC1) improves whole body glucose tolerance and insulin sensitivity in pregnant mice. Biochim Biophys Acta Mol Basis Dis. 2019;1865:6
94.
Cao H, Chen X, Yang Y, Storm DR. Disruption of type 3 adenylyl cyclase expression in the hypothalamus leads to obesity. Integr Obes Diabetes. 2016;2:225-228.
95.
Appaiahgari MB, Vrati S. Adenoviruses as gene/vaccine delivery vectors: promises and pitfalls. Expert Opin Biol Ther. 2015;15:337-351.
96.
Sinnayah P, Lindley TE, Staber PD, Davidson BL, Cassell MD, Davisson RL. Targeted viral delivery of Cre recombinase induces conditional gene deletion in cardiovascular circuits of the mouse brain. Physiol Genomics. 2004;18:25-32.
97.
Xiang D, Tao L, Li Z. Modeling Breast Cancer via an Intraductal Injection of Cre-expressing Adenovirus into the Mouse Mammary Gland. J Vis Exp. 2019.
98.
Tao L, van Bragt MP, Laudadio E, Li Z. Lineage tracing of mammary epithelial cells using cell-type-specific cre-expressing adenoviruses. Stem Cell Reports. 2014;2:770-779.
99.
Penny GM, Cochran RB, Pihlajoki M, Kyronlahti A, Schrade A, Hakkinen M, et al. Probing GATA factor function in mouse Leydig cells via testicular injection of adenoviral vectors. Reproduction. 2017;154:455-467.
100.
Yu PB, Deng DY, Lai CS, Hong CC, Cuny GD, Bouxsein ML, et al. BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat Med. 2008;14:1363-1369.
101.
Kobayashi N, Noguchi H, Westerman KA, Matsumura T, Watanabe T, Totsugawa T, et al. Efficient Cre/loxP site-specific recombination in a HepG2 human liver cell line. Cell Transplant. 2000;9:737-742.
102.
Gan Y, Zhang Y, Digirolamo DJ, Jiang J, Wang X, Cao X, et al. Deletion of IGF-I receptor (IGF-IR) in primary osteoblasts reduces GH-induced STAT5 signaling. Mol Endocrinol. 2010;24:644-656.
103.
Cao Y, Pan T, Chen X, Wu J, Guo N, Wang B. EP4 knockdown alleviates glomerulosclerosis through Smad and MAPK pathways in mesangial cells. Mol Med Rep. 2018;18:5141-5150.
104.
Gan Y, Paterson AJ, Zhang Y, Jiang J, Frank SJ. Functional collaboration of insulin-like growth factor-1 receptor (IGF-1R), but not insulin receptor (IR), with acute GH signaling in mouse calvarial cells. Endocrinology. 2014;155:1000-1009.
105.
Carofino BL, Justice MJ. Tissue-Specific Regulation of Oncogene Expression Using Cre-Inducible ROSA26 Knock-In Transgenic Mice. Curr Protoc Mouse Biol. 2015;5:187-204.
106.
Albert K, Voutilainen MH, Domanskyi A, Piepponen TP, Ahola S, Tuominen RK, et al. Downregulation of tyrosine hydroxylase phenotype after AAV injection above substantia nigra: Caution in experimental models of Parkinson's disease. J Neurosci Res. 2019;97:346-3
107.
Depuy SD, Kanbar R, Coates MB, Stornetta RL, Guyenet PG. Control of breathing by raphe obscurus serotonergic neurons in mice. J Neurosci. 2011;31:1981-1990.
108.
Dugue GP, Akemann W, Knopfel T. A comprehensive concept of optogenetics. Prog Brain Res. 2012;196:1-28.
109.
Kim B, Lin MZ. Optobiology: optical control of biological processes via protein engineering. Biochem Soc Trans. 2013;41:1183-1188.
110.
DePuy SD, Stornetta RL, Bochorishvili G, Deisseroth K, Witten I, Coates M, et al. Glutamatergic neurotransmission between the C1 neurons and the parasympathetic preganglionic neurons of the dorsal motor nucleus of the vagus. J Neurosci. 2013;33:1486-1497.
- GENEMEDI
- Email: [email protected] [email protected]
- Telephone: +86-21-50478399 Fax: 86-21-50478399
- Privacy Policy
- TECHNICAL SUPPORT
- Product FAQs
- Technical manuals
- Publications
- Email: [email protected]









 Facebook
Facebook LinkedIn
LinkedIn Twitter
Twitter